Available in a git repository.
Repository: depgraph
Browsable repository: depgraph
Author: W. Trevor King
This set of scripts scans a Python file for module dependencies, and generates code for Graphviz dot. See call graph for a script that generates dependency graphs within a module.
Originally by Toby Dickenson, the scripts are good for giving you the "long view" on how your project is organized. I've altered the originals a bit.
My py2depgraph.py also prints out path information for each module, so that you can show or hide module nodes based on those paths. For example, ignoring system modules to focus on your additions.
My depgraph2dot.py has an added method
depgraph2dot.invalidpath(module_name, path) that you can override to
determine the paths you like.
I've also added the depgraph2dot methods
is_Cext(module_name, path),Cext_depends(module_name, path),Cext_edge_attributes(module_name, Cext_name),Cext_node_attributes(Cext_name), andCext_depcolor(Cext_name)
to detect and configure the shared C libraries extending Python, so you know where to look for the code your Python depends on.
Available in a git repository.
Repository: unfold_protein
Browsable repository: unfold_protein
Author: W. Trevor King
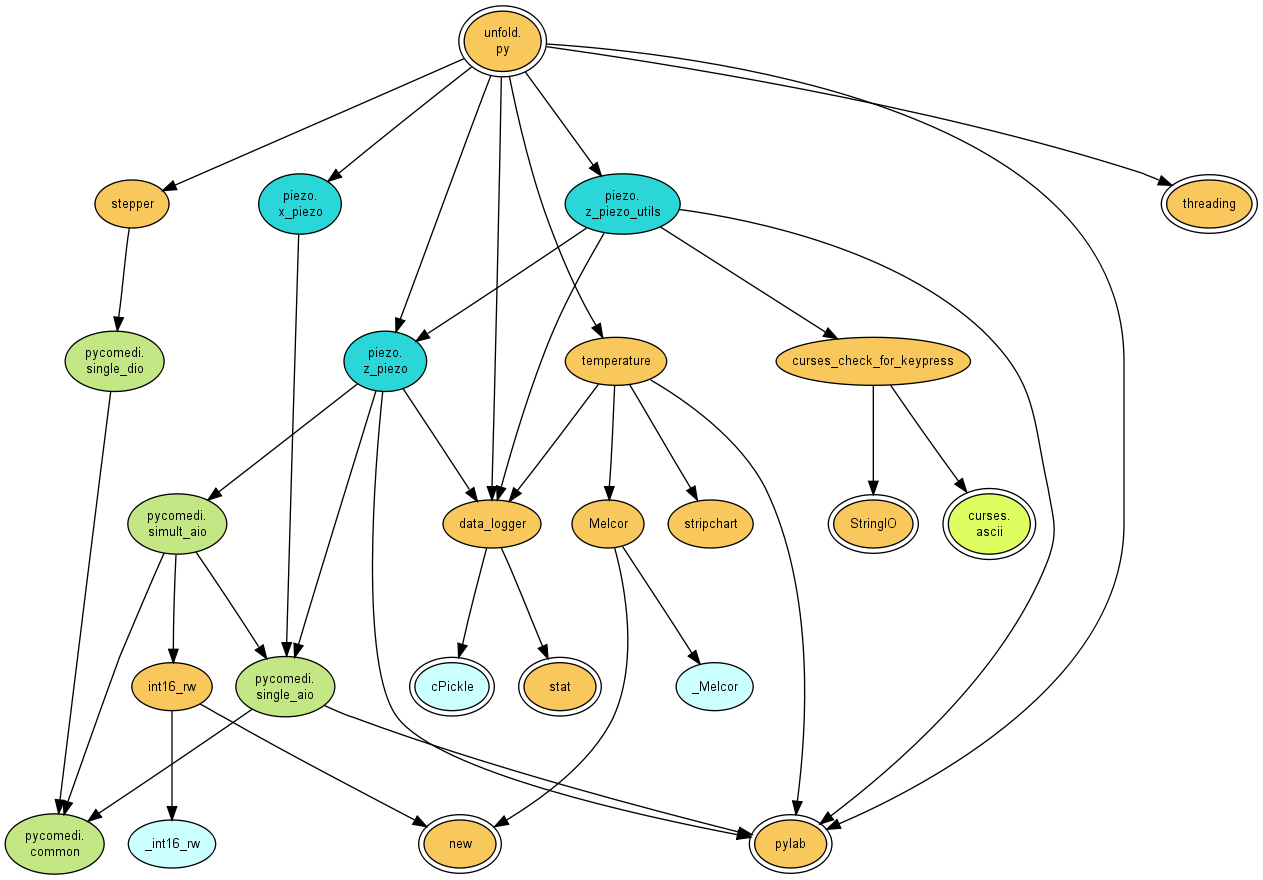
As example, here is the depgraph of my protein unfolding program unfold.py
$ python py2depgraph.py unfold.py | python depgraph2dot.py | dot -T png -o depgraph.png
with standard python modules in orange,
python packages in hash-based colors,
and C extension modules in light blue.
Again, these are only the files containing either wking or comedi
in their paths.