My publications.
My current work for Prof. Yang involves unfolding proteins using an Atomic Force Microscope (AFM) at different temperatures to estimate the roughness of their free energy landscape. For a brief overview of force spectroscopy, see my two second summary. For more detail, you can look at some of my papers.
I've written some experimental control software, a Monte Carlo simulation program (sawsim) for analyzing the unfolding sawtooth curves, as well as software for calibrating AFM cantilevers via the thermal tune method (calibcant), scaling the unfolding data, selecting good curves, and fitting those curves with wormlike chains (Hooke).
Along the way I've had to learn way too much about the internal workings of our MultiMode AFM.
I've also posted my notes for our Debian cluster.
Available in a git repository.
Repository: thesis
Browsable repository: thesis
Author: W. Trevor King
The source for my Ph.D. thesis (in progress). A draft is compiled after each commit. I use the drexel-thesis class, which I also maintain.
Exciting features: SCons build; Asymptote/asyfig, PGF, and PyMOL figures.
Available in a git repository.
Repository: sawsim
Browsable repository: sawsim
Author: W. Trevor King
Introduction
My thesis project investigates protein unfolding via the experimental technique of force spectroscopy. In force spectroscopy, we mechanically stretch chains of proteins, usually by pulling one end of the chain away from a surface with an AFM.
For velocity clamp experiments (the simplest to carry out experimentally), the experiments produce "sawtooth" force-displacement curves. As the protein stretches, the tension increases. At some point, a protein domain unfolds, increasing the total length of the chain and relaxing the tension. As we continue to stretch the protein, we see a series of unfolding peaks. The GPLed program Hooke analyzes the sawtooth curves and extracts lists of unfolding forces.
Lists of unfolding forces are not particularly interesting by themselves. The most common approach for extracting some physical insights from the unfolding curves is to take a guess at an explanatory model and check the predicted behavior of the model against the measured behavior of the protein. If the model does a good job of explaining the protein behavior, it might be what's actually going on behind the scenes. Sawsim is my (published!) tool for simulating force spectroscopy experiments and matching the simulations to experimental results.
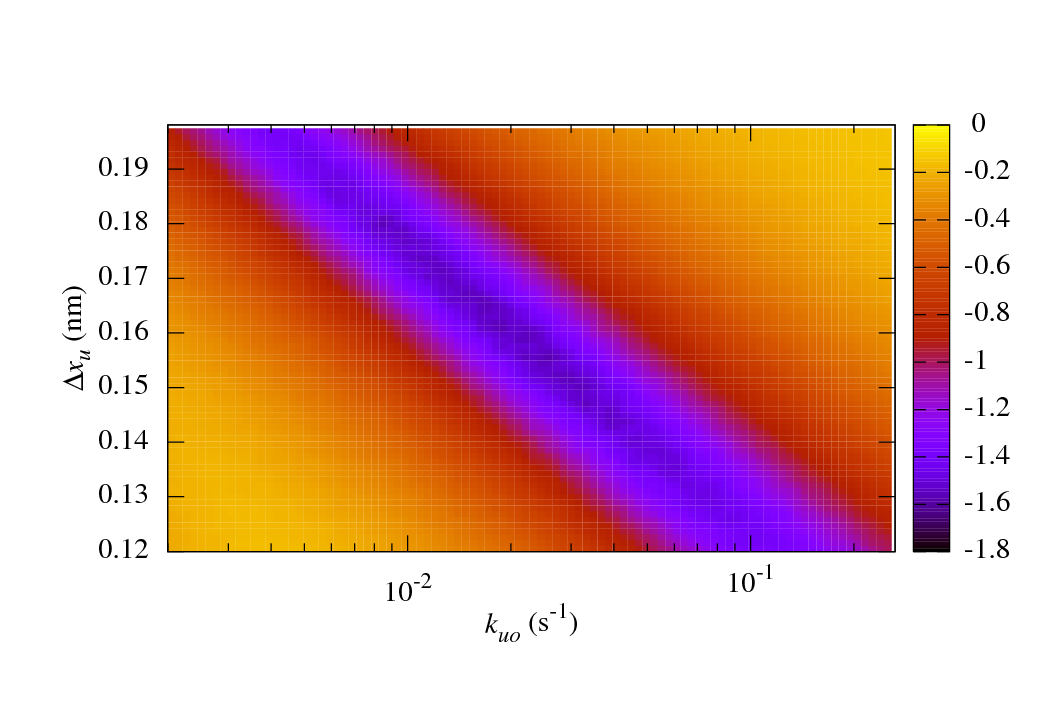
The main benefits of sawsim are its ability to simulate systems with arbitrary numbers of states (see the manual) and to easily compare the simulated data with experimental values. The following figure shows a long valley of reasonable fits to some ubiquitin unfolding data. See the IJBM paper (linked above) for more details.
Getting started
Sawsim should run anywhere you have a C compiler and Python 2.5+. I've tested it on Gentoo and Debian, and I've got an ebuild in my Gentoo overlay. It should also run fine on Windows, etc., but I don't have access to any Windows boxes with a C compiler, so I haven't tested that (email me if you have access to such a machine and want to try installing Sawsim).
See the README, manual, and PyPI page for more details.